DocGenome: An Open Large-scale Scientific Document Benchmark for Training Next-generation Large Models
Published in 500K scientific documents including 6.5M pages with 13 attributes of component units, 6 logical relationships between units, and 7 document-oriented tasks, 2024
Overall
This blog will introduce our proposed large-scale scientific dataset, DocGenome, which is constructed using our custom auto-labeling pipeline, DocParser.
DocParser is a cutting-edge auto-labeling pipeline, which can generate both attribute information of component units and logical relationships between units by auto-annotating and structuring a large amount of unlabeled arXiv papers, with four stages:
- 1) data preprocessing,
- 2) unit segmentation,
- 3) attribute assignment and relation retrieval
- 4) color rendering
By DocParser, we construct the DocGenome by auto-annotating and structuring unlabeled arXiv papers, and it features four characteristics:
- 1) Completeness: It is the first dataset to structure data from all modalities including 13 layout attributes along with their LaTeX source codes.
- 2) Logicality: It provides 6 logical relationships between different entities within each scientific document.
- 3) Diversity: It covers various document-oriented tasks, including document classification, visual grounding, document layout detection, document transformation, open-ended single-page QA and multi-page QA.
- 4) Correctness: It undergoes rigorous quality control checks conducted by a specialized team.
Besides, based on DocGenome, we conduct extensive experiments to demonstrate the advantages of DocGenome and objectively evaluate the performance of current large models on our benchmark.


Release
- [2024/6/10] 🔥 Our paper entitled “DocGenome: An Open Large-scale Scientific Document Benchmark for Training and Testing Multi-modal Large Models” has been released in arXiv Link
- [2024/6/6] 🔥 We have released the DocGenome benchmark, includes 8 subsets as follows:
DocGenome Benchmark Introduction
| Datasets | # Discipline | # Category of Units | # Pages in Train-set | # Pages in Test-set | # Task | # Used Metric | Publication | Entity Relations |
|---|---|---|---|---|---|---|---|---|
| DocVQA | - | N/A | 11K | 1K | 1 | 2 | 1960-2000 | ❎ |
| DocLayNet | - | 11 | 80K | 8K | 1 | 1 | - | ❎ |
| DocBank | - | 13 | 0.45M | 50K | 3 | 1 | 2014-2018 | ❎ |
| PubLayNet | - | 5 | 0.34M | 12K | 1 | 1 | - | ❎ |
| VRDU | - | 10 | 7K | 3K | 3 | 1 | - | ❎ |
| DUDE | - | N/A | 20K | 6K | 3 | 3 | 1860-2022 | ❎ |
| D^4LA | - | 27 | 8K | 2K | 1 | 3 | - | ❎ |
| Fox Benchmark | - | 5 | N/A (No train-set) | 0.2K | 3 | 5 | - | ❎ |
| ArXivCap | 32 | N/A | 6.4M* | N/A | 4 | 3 | - | ❎ |
| DocGenome (ours) | 153 | 13 | 6.8M | 9K | 7 | 7 | 2007-2022 | ✅ |
👇🏻DocGenome-train Download
We provide 8 subsets of DocGenome-train for downloading:
Data Download
- [docgenome-train-000.tar.gz]() - [docgenome-train-001.tar.gz]() - [docgenome-train-002.tar.gz]() - [docgenome-train-003.tar.gz]() - [docgenome-train-004.tar.gz]() - [docgenome-train-005.tar.gz]() - [docgenome-train-006.tar.gz]() - [docgenome-train-007.tar.gz]()Definition of relationships between component units
DocGenome contains 4 level relation types and 2 cite relation types, as shown in the following table:
| Name | Description | Example | |——————————|——————————————————————|—————————————————————————-| | Identical | Two blocks share the same source code. | Cross-column text; Cross-page text. | | Title adjacen | The two titles are adjacent. | (\textbackslash section{introduction}, \textbackslash section{method}) | | Subordinate | One block is a subclass of another block. | (\textbackslash section{introduction}, paragraph within Introduction) | | Non-title adjacent | The two text or equation blocks are adjacent. | (Paragraph 1, Paragraph 2) | | Explicitly-referred | One block refers to another block via footnote, reference, etc. | (As shown in \textbackslash ref{Fig: 5} …, Figure 5) | | Implicitly-referred | The caption block refers to the corresponding float environment. | (Table Caption 1, Table 1)
</details>
Attribute of component units
DocGenome has 13 attributes of component units, which can be categorized into two classes
- 1) Fixed-form units, including Text, Title, Abstract, etc., which are characterized by sequential reading and hierarchical relationships readily discernible from the list obtained in Stage-two of the designed DocParser.
- 2) Floating-form units, including Table, Figure, etc., which establish directional references to fixed-form units through commands like \ref and \label.
| Index | Category | Notes |
|---|---|---|
| 0 | Algorithm | |
| 1 | Caption | Titles of Images, Tables, and Algorithms |
| 2 | Equation | |
| 3 | Figure | |
| 4 | Footnote | |
| 5 | List | |
| 7 | Table | |
| 8 | Text | |
| 9 | Text-EQ | Text block with inline equations |
| 10 | Title | Section titles |
| 12 | PaperTitle | |
| 13 | Code | |
| 14 | Abstract |
Note that we do not use the “others” category and the “reference” category, and their indices are 6 and 11, respectively.
Types of disciplines
Page distribution of DocGenome. 20\% of documents are five pages or fewer, 50\% are ten pages or fewer, and 80\% are nineteen pages or fewer.
Page Distribution

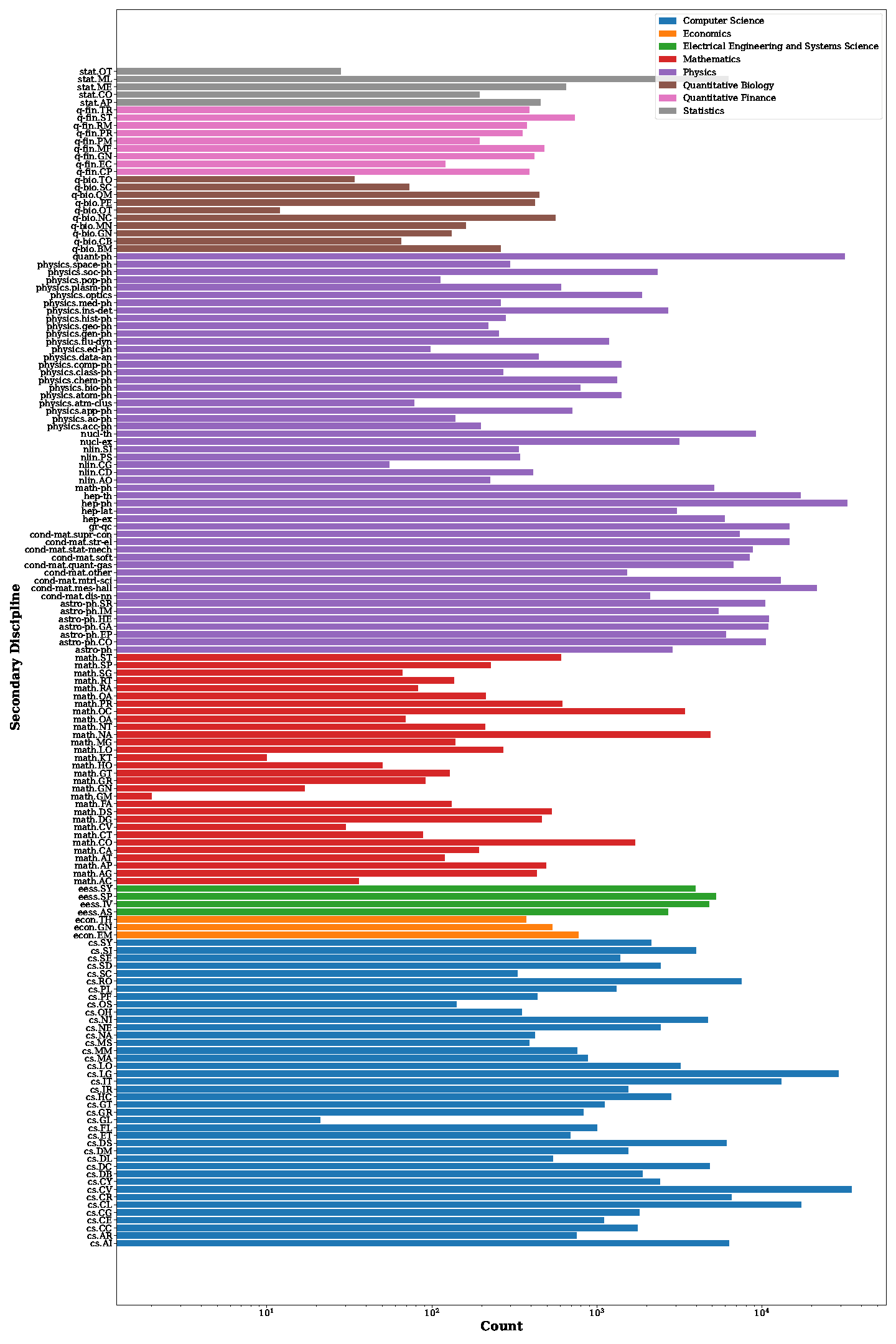
Distribution of secondary disciplines in our DocGenome. The count on the x-axis represents the number of documents, and documents from the same primary discipline are marked with the same color.
Discipline Distribution
DocParser: A Cutting-edge Auto-labeling Pipeline
Schematic of the designed DocParser pipeline for automated document annotation The process is divided into four distinct stages:
- 1) Data Preprocessing,
- 2) Unit Segmentation,
- 3) Attribute Assignment and Relation Retrieval,
- 4) Color Rendering.
DocParser can convert LaTeX source code of a complete document into annotations for component units with source-code, attributes, relationships and bounding box, as well as a rendered PNG of the entire document.

Visualizations
Visual examples of document-oriented tasks in DocGenome

Citation
If you find our work useful in your research, please consider citing Fox:
@article{,
}
